Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
Por um escritor misterioso
Last updated 09 abril 2025


Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates
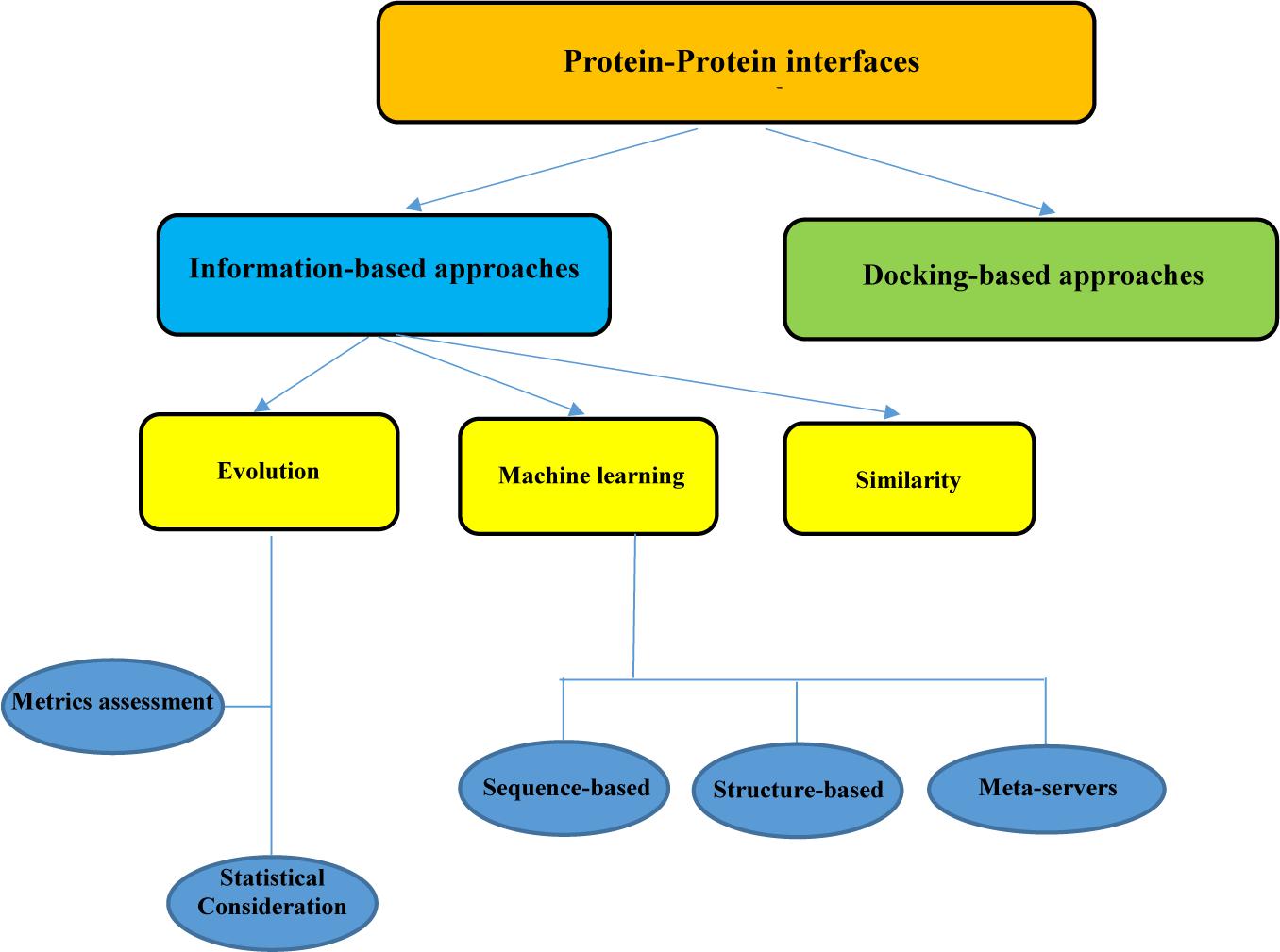
Frontiers In silico Approaches for the Design and Optimization of Interfering Peptides Against Protein–Protein Interactions
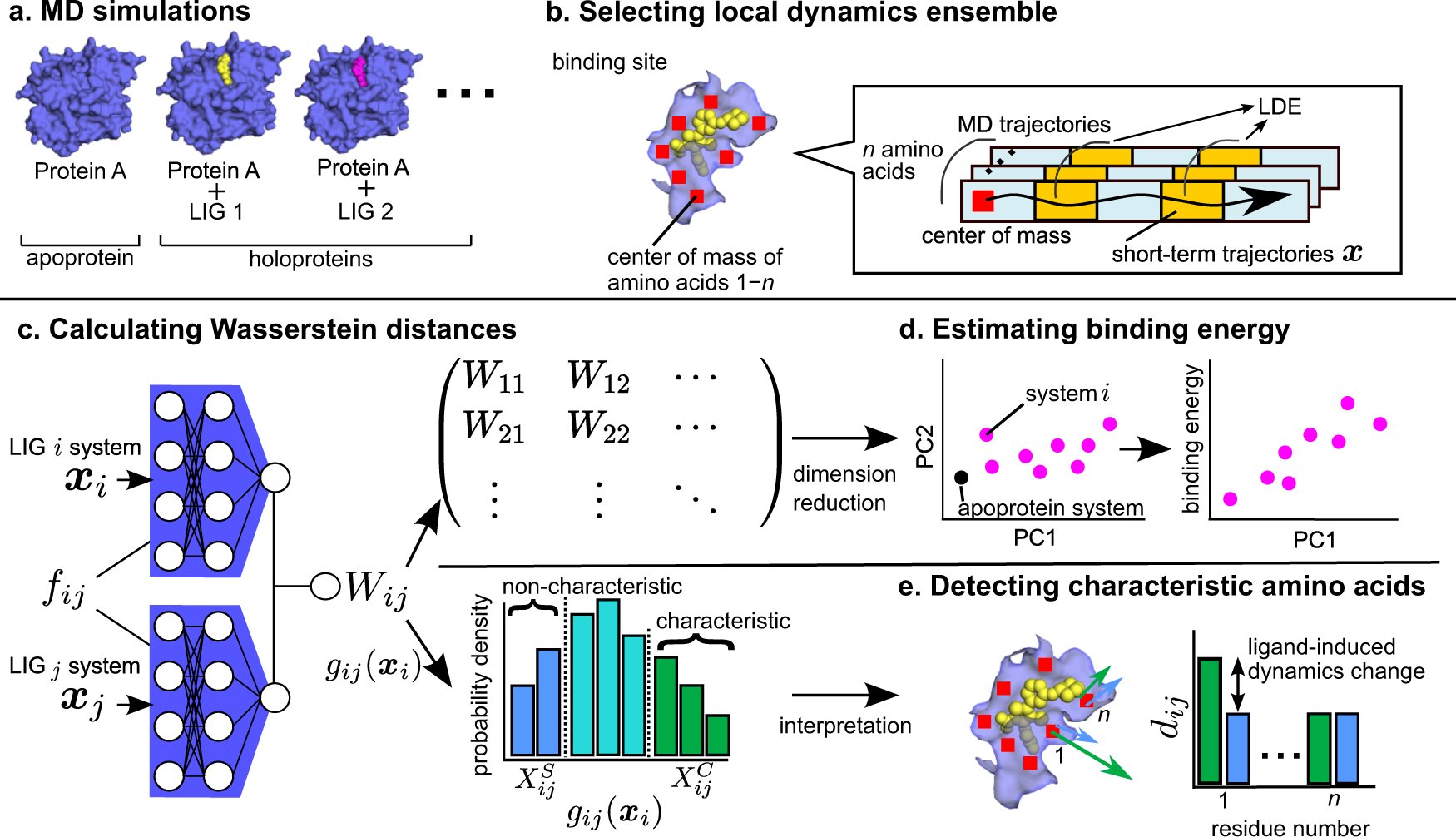
Differences in ligand-induced protein dynamics extracted from an unsupervised deep learning approach correlate with protein–ligand binding affinities
The use of machine learning modeling, virtual screening, molecular docking, and molecular dynamics simulations to identify potential VEGFR2 kinase inhibitors

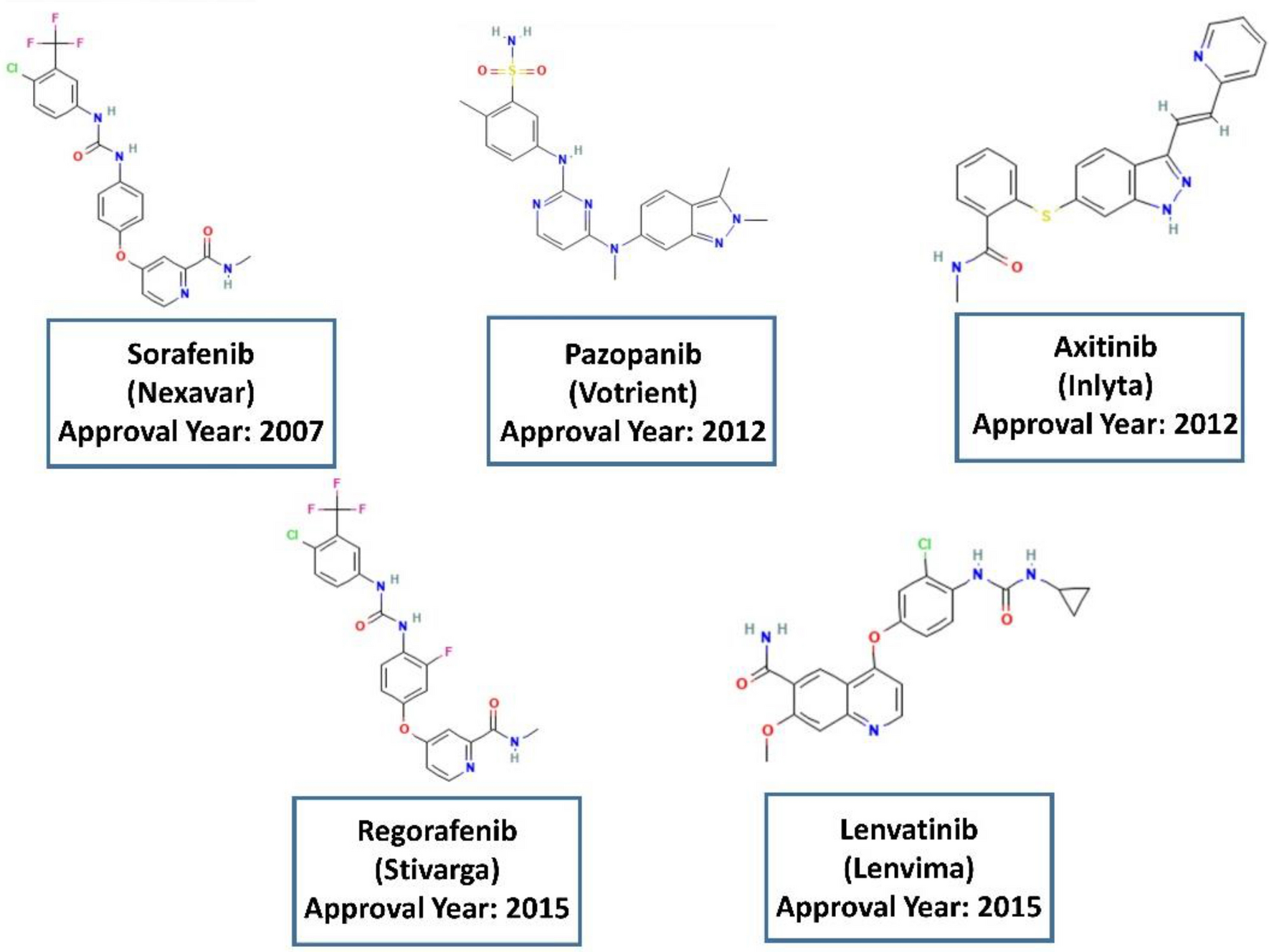
Shown are chemical structures of the 6 incorrectly predicted P-gp
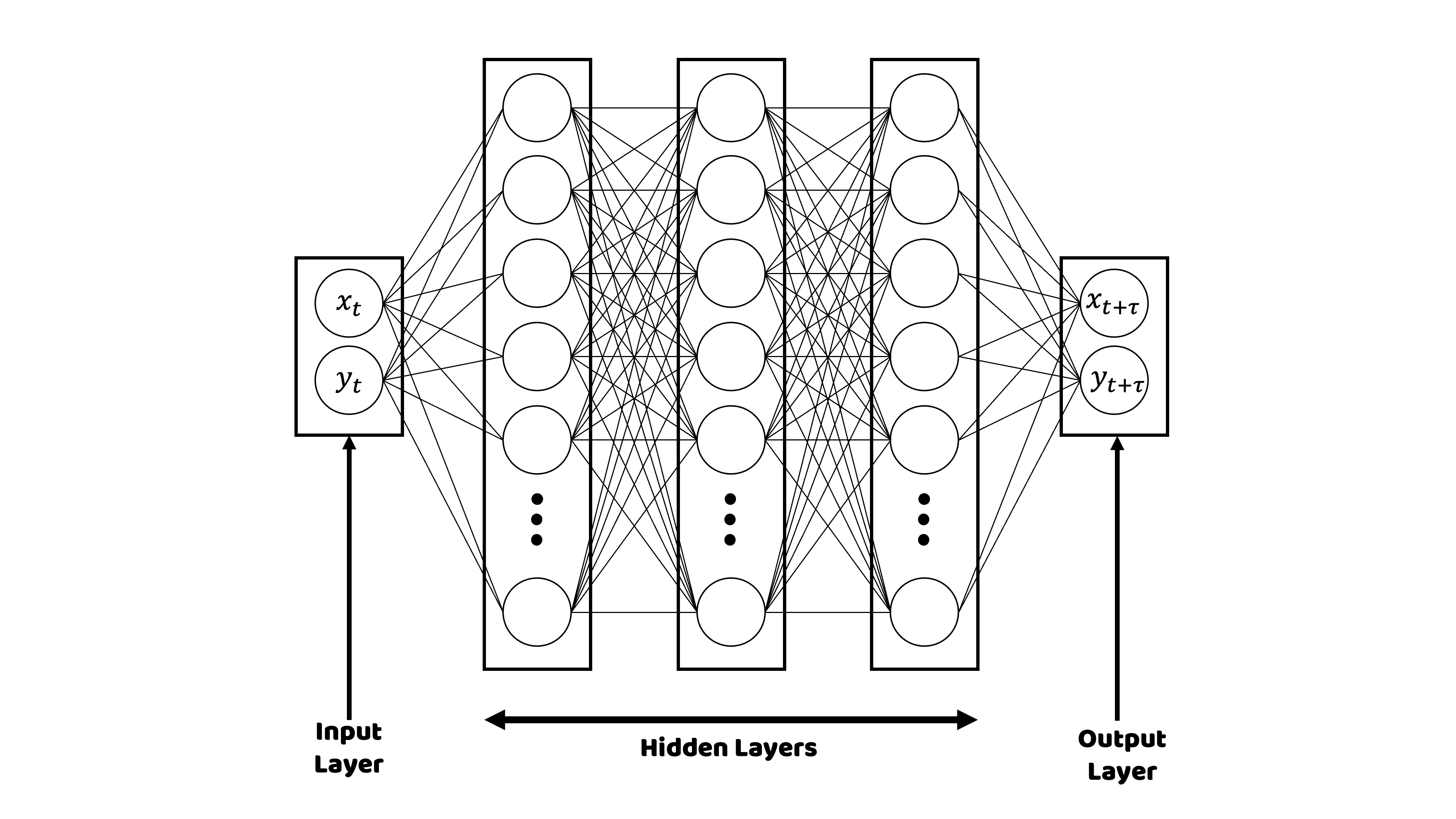
BioSimLab - Research

Multisite model for P-glycoprotein drug binding. MOLCAD representation

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Combining Machine Learning and Molecular Dynamics to Predict P-Glycoprotein Substrates

Deep learning-based molecular dynamics simulation for structure-based drug design against SARS-CoV-2 - ScienceDirect

Effect of the Force Field on Molecular Dynamics Simulations of the Multidrug Efflux Protein P-Glycoprotein

Prediction and characterization of P-glycoprotein substrates potentially bound to different sites by emerging chemical pattern and hierarchical cluster analysis - ScienceDirect
Recomendado para você
-
 Brain Test Level 56 How many eggs are there in 202309 abril 2025
Brain Test Level 56 How many eggs are there in 202309 abril 2025 -
Erase Puzzle for Android - Download the APK from Uptodown09 abril 2025
-
 Guess the Sneakers! Kicks Quiz for Sneakerheads by Tapgang Apps and Games, Inc.09 abril 2025
Guess the Sneakers! Kicks Quiz for Sneakerheads by Tapgang Apps and Games, Inc.09 abril 2025 -
 Vessel Part 1-(page 1&2)09 abril 2025
Vessel Part 1-(page 1&2)09 abril 2025 -
MTVBaseAfrica09 abril 2025
-
 A Genetic Screen Implicates miRNA-372 and miRNA-373 As Oncogenes in Testicular Germ Cell Tumors: Cell09 abril 2025
A Genetic Screen Implicates miRNA-372 and miRNA-373 As Oncogenes in Testicular Germ Cell Tumors: Cell09 abril 2025 -
 Autonomy and the principle of respect for autonomy.09 abril 2025
Autonomy and the principle of respect for autonomy.09 abril 2025 -
Arithmetic & Geometric Sequences, 154 plays09 abril 2025
-
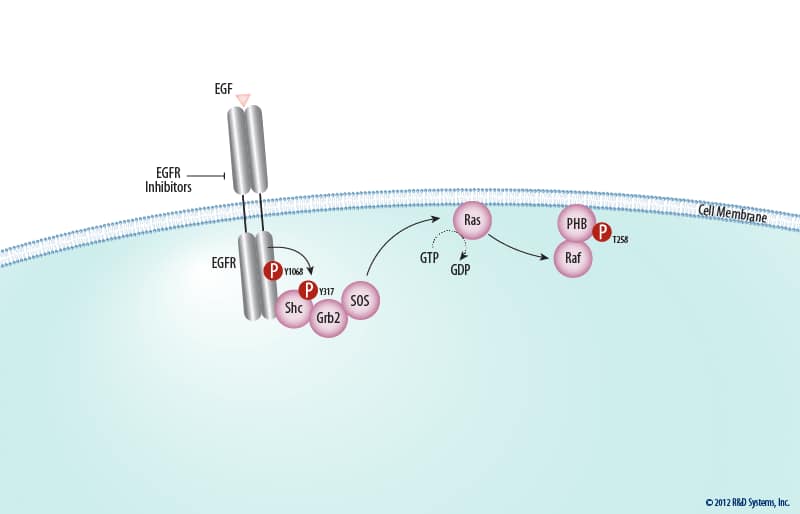 The ERK Signal Transduction Pathway: R&D Systems09 abril 2025
The ERK Signal Transduction Pathway: R&D Systems09 abril 2025 -
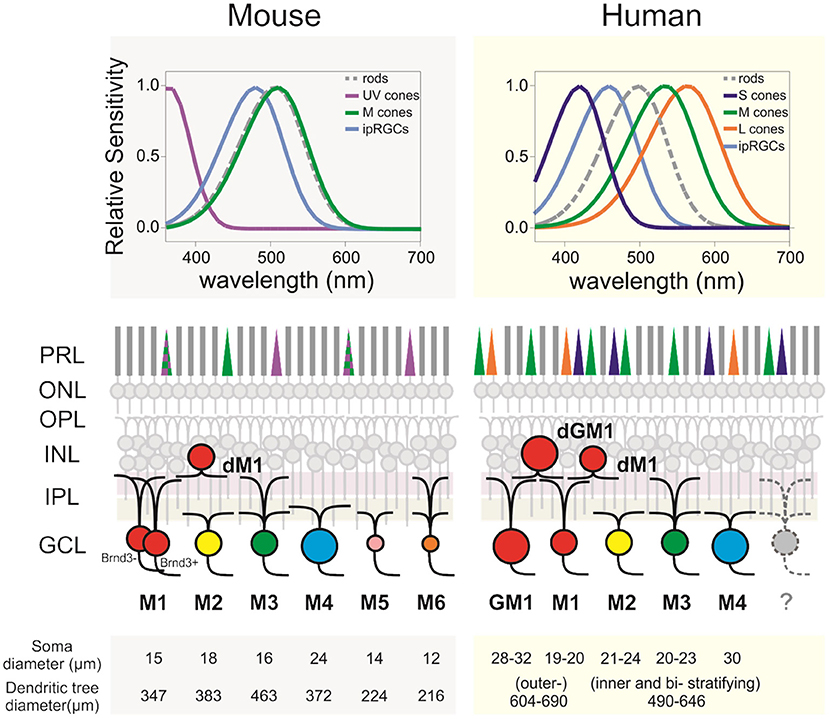 Frontiers Intrinsically Photosensitive Retinal Ganglion Cells of the Human Retina09 abril 2025
Frontiers Intrinsically Photosensitive Retinal Ganglion Cells of the Human Retina09 abril 2025
você pode gostar
-
 We Need More Games Like The Outer Wilds09 abril 2025
We Need More Games Like The Outer Wilds09 abril 2025 -
 Grand Theft Auto VI leak followed by an official trailer with a twist: A release date of 202509 abril 2025
Grand Theft Auto VI leak followed by an official trailer with a twist: A release date of 202509 abril 2025 -
 ValueAct aumenta participação na Disney, diz agência09 abril 2025
ValueAct aumenta participação na Disney, diz agência09 abril 2025 -
 The Last of Us Part I Latest 1.0.4 PC Update Offers CPU/GPU Optimization, Steam Deck Fix and More09 abril 2025
The Last of Us Part I Latest 1.0.4 PC Update Offers CPU/GPU Optimization, Steam Deck Fix and More09 abril 2025 -
 Gears 5 - Hivebusters DLC - Gameplay Walkthrough (FULL CAMPAIGN)09 abril 2025
Gears 5 - Hivebusters DLC - Gameplay Walkthrough (FULL CAMPAIGN)09 abril 2025 -
 What Does Gene Munster Have to Say About an Apple Inc. (AAPL09 abril 2025
What Does Gene Munster Have to Say About an Apple Inc. (AAPL09 abril 2025 -
 Novos detalhes de Pokémon Origins09 abril 2025
Novos detalhes de Pokémon Origins09 abril 2025 -
 TSV Sementes - product09 abril 2025
TSV Sementes - product09 abril 2025 -
 Is Tim Cook Right? Are Apps the Future of TV? NCTA — The Internet & Television Association09 abril 2025
Is Tim Cook Right? Are Apps the Future of TV? NCTA — The Internet & Television Association09 abril 2025 -
 Unveiling the Metaverse 2023-2024: From Sci-Fi Dream to Digital09 abril 2025
Unveiling the Metaverse 2023-2024: From Sci-Fi Dream to Digital09 abril 2025