Evaluation of taxonomic classification and profiling methods for long-read shotgun metagenomic sequencing datasets, BMC Bioinformatics
Por um escritor misterioso
Last updated 11 abril 2025
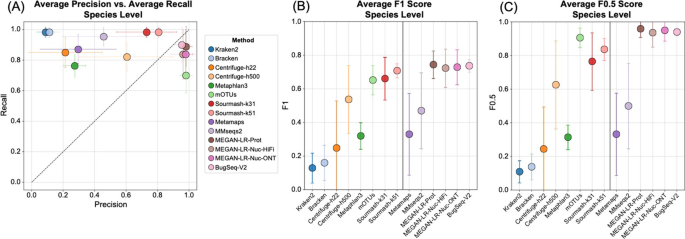
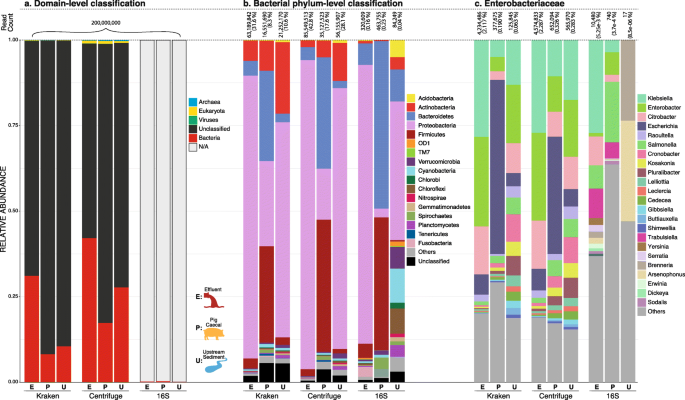
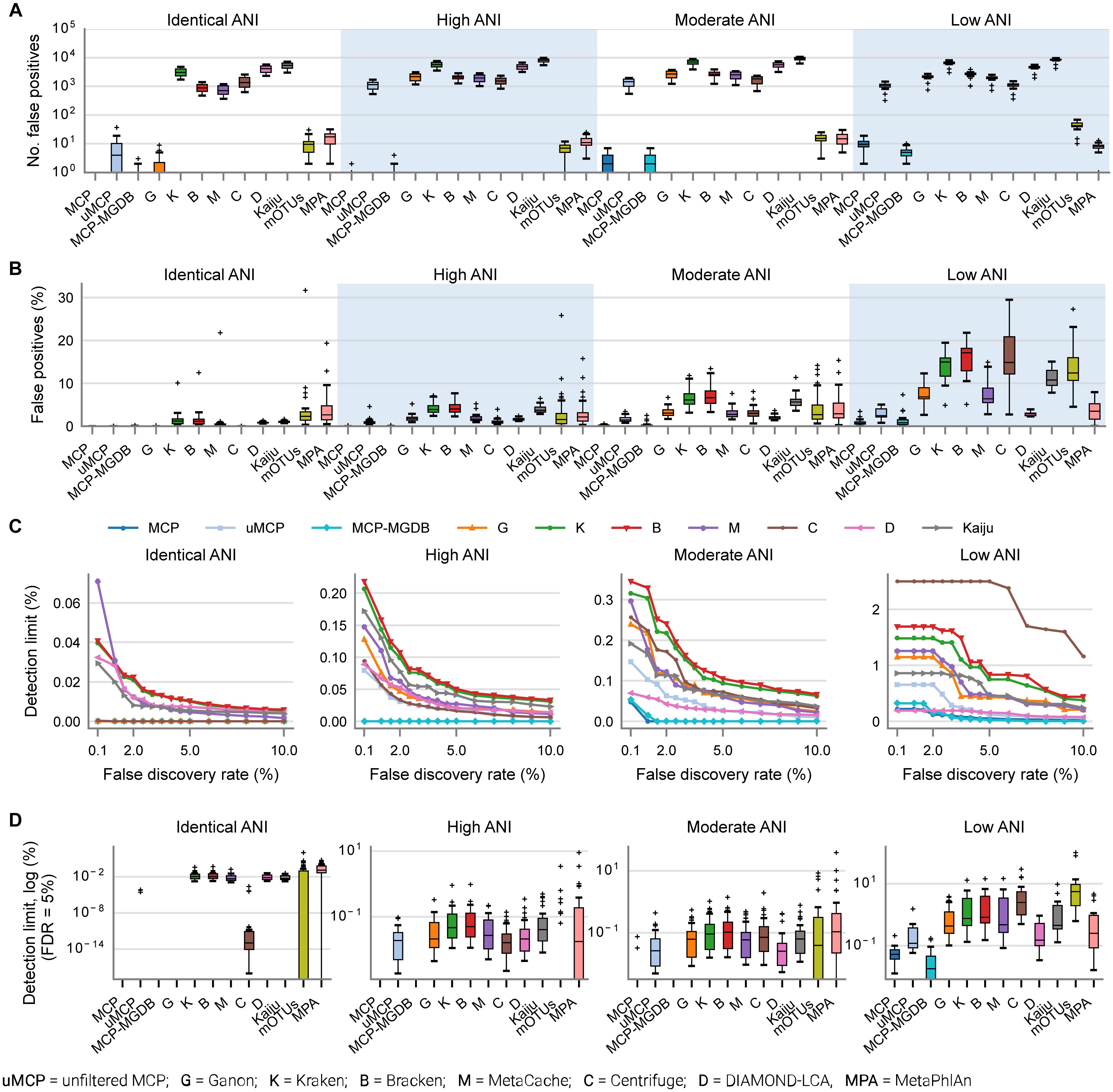
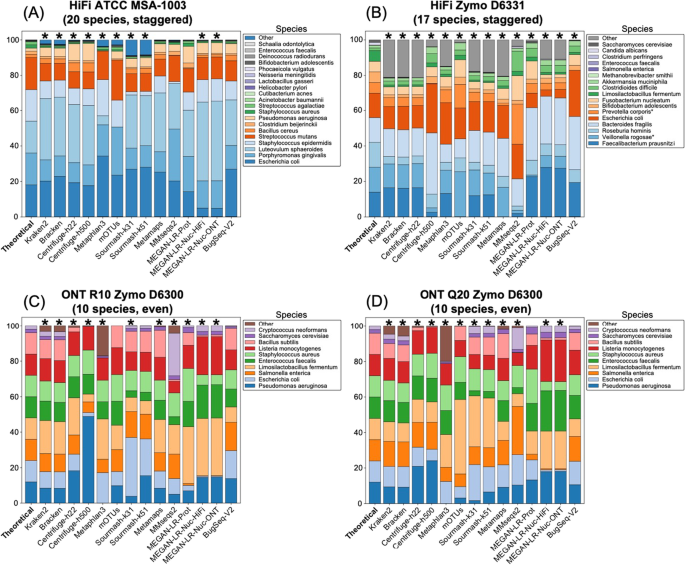
Background Long-read shotgun metagenomic sequencing is gaining in popularity and offers many advantages over short-read sequencing. The higher information content in long reads is useful for a variety of metagenomics analyses, including taxonomic classification and profiling. The development of long-read specific tools for taxonomic classification is accelerating, yet there is a lack of information regarding their relative performance. Here, we perform a critical benchmarking study using 11 methods, including five methods designed specifically for long reads. We applied these tools to several mock community datasets generated using Pacific Biosciences (PacBio) HiFi or Oxford Nanopore Technology sequencing, and evaluated their performance based on read utilization, detection metrics, and relative abundance estimates. Results Our results show that long-read classifiers generally performed best. Several short-read classification and profiling methods produced many false positives (particularly at lower abundances), required heavy filtering to achieve acceptable precision (at the cost of reduced recall), and produced inaccurate abundance estimates. By contrast, two long-read methods (BugSeq, MEGAN-LR & DIAMOND) and one generalized method (sourmash) displayed high precision and recall without any filtering required. Furthermore, in the PacBio HiFi datasets these methods detected all species down to the 0.1% abundance level with high precision. Some long-read methods, such as MetaMaps and MMseqs2, required moderate filtering to reduce false positives to resemble the precision and recall of the top-performing methods. We found read quality affected performance for methods relying on protein prediction or exact k-mer matching, and these methods performed better with PacBio HiFi datasets. We also found that long-read datasets with a large proportion of shorter reads (< 2 kb length) resulted in lower precision and worse abundance estimates, relative to length-filtered datasets. Finally, for classification methods, we found that the long-read datasets produced significantly better results than short-read datasets, demonstrating clear advantages for long-read metagenomic sequencing. Conclusions Our critical assessment of available methods provides best-practice recommendations for current research using long reads and establishes a baseline for future benchmarking studies.

Atcc Msa 1003 (ATCC), Bioz
The impact of sequencing depth on the inferred taxonomic

Current concepts, advances, and challenges in deciphering the
Frontiers Evaluation of the Microba Community Profiler for

Benchmarking Metagenomics Tools for Taxonomic Classification

Evaluation of taxonomic classification and profiling methods for

Evaluation of taxonomic classification and profiling methods for
Evaluation of taxonomic classification and profiling methods for
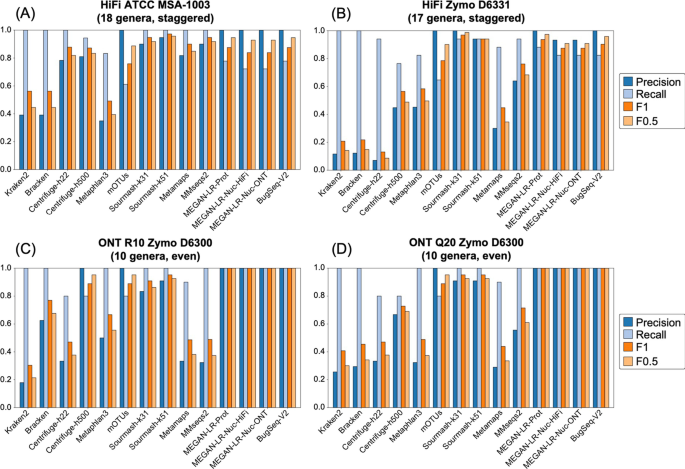
Evaluation of taxonomic classification and profiling methods for

Evaluation of taxonomic classification and profiling methods for

Full article: Recommendations for the use of metagenomics for
Recomendado para você
-
 > The Official Murder Mystery 2's Value List11 abril 2025
> The Official Murder Mystery 2's Value List11 abril 2025 -
flames godly mm2 value|TikTok Search11 abril 2025
-
splitter mm2 value|TikTok Search11 abril 2025
-
Flexible Magnetic Field Nanosensors for Wearable Electronics: A Review11 abril 2025
-
 Trading this for Spectre set, I'm top and use supreme : r/MurderMystery211 abril 2025
Trading this for Spectre set, I'm top and use supreme : r/MurderMystery211 abril 2025 -
 Area-Selective Etching of Poly(methyl methacrylate) Films by11 abril 2025
Area-Selective Etching of Poly(methyl methacrylate) Films by11 abril 2025 -
 Ez trading on MM2 : r/MurderMystery211 abril 2025
Ez trading on MM2 : r/MurderMystery211 abril 2025 -
 FRS concept using an Nd:YAG laser and molecular iodine filter11 abril 2025
FRS concept using an Nd:YAG laser and molecular iodine filter11 abril 2025 -
 Phosphogermanate Crystal: A New Ultraviolet–Infrared Nonlinear11 abril 2025
Phosphogermanate Crystal: A New Ultraviolet–Infrared Nonlinear11 abril 2025 -
 40 Feet 20 AWG High Temperature PTFE Silver Plated Wire 0.5mm211 abril 2025
40 Feet 20 AWG High Temperature PTFE Silver Plated Wire 0.5mm211 abril 2025
você pode gostar
-
 Samsung expande oferta de streaming de games para mais modelos de TVs 4K – Samsung Newsroom Brasil11 abril 2025
Samsung expande oferta de streaming de games para mais modelos de TVs 4K – Samsung Newsroom Brasil11 abril 2025 -
 A Nova Temporada Americana - Globo Rural Edição 413 - Missão Mulheres do Agro11 abril 2025
A Nova Temporada Americana - Globo Rural Edição 413 - Missão Mulheres do Agro11 abril 2025 -
 Balder Hringhorni/Kamigami no Asobi by LaiKa86 on DeviantArt11 abril 2025
Balder Hringhorni/Kamigami no Asobi by LaiKa86 on DeviantArt11 abril 2025 -
Fast Game - Booster - Apps on Google Play11 abril 2025
-
 Fundo Elfo De Desenho Animado Fundo, Páginas Para Colorir De Fantasia, Imagem De Um Elfo Para Colorir, Duende Imagem de plano de fundo para download gratuito11 abril 2025
Fundo Elfo De Desenho Animado Fundo, Páginas Para Colorir De Fantasia, Imagem De Um Elfo Para Colorir, Duende Imagem de plano de fundo para download gratuito11 abril 2025 -
 Camisa Pré Jogo do Flamengo 23 adidas - Infantil11 abril 2025
Camisa Pré Jogo do Flamengo 23 adidas - Infantil11 abril 2025 -
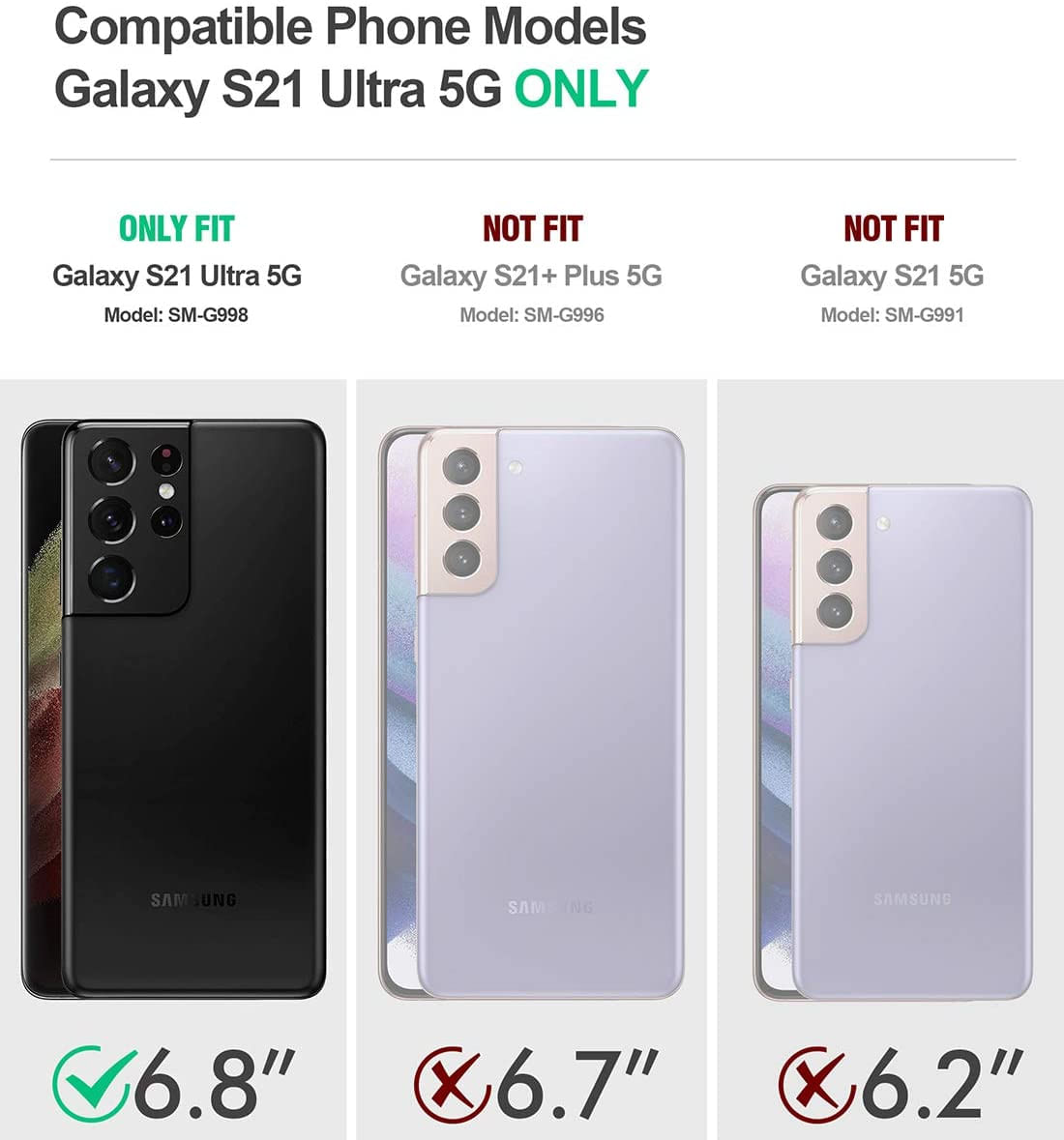 Galaxy s21 ultra 5g em promoção11 abril 2025
Galaxy s21 ultra 5g em promoção11 abril 2025 -
 Compra online de Jogo de tabuleiro de futebol de mesa com 10 bolas, brinquedos interativos pai-filho, mini bola de jogo de mesa, brinquedos de futebol, jogos competitivos11 abril 2025
Compra online de Jogo de tabuleiro de futebol de mesa com 10 bolas, brinquedos interativos pai-filho, mini bola de jogo de mesa, brinquedos de futebol, jogos competitivos11 abril 2025 -
 Freefire logo vector, Freefire icon free vector 20190574 Vector11 abril 2025
Freefire logo vector, Freefire icon free vector 20190574 Vector11 abril 2025 -
 Inter, Correa salta il derby col Milan. Le probabili formazioni - la Repubblica11 abril 2025
Inter, Correa salta il derby col Milan. Le probabili formazioni - la Repubblica11 abril 2025


